Exploring the PlanktoScope as a tool for summer HAB monitoring
The Cohen lab planktoscope!
By Mary Reed, SkIO Summer Intern 2025
What is the PlanktoScope?
The PlanktoScope is an open-source imaging system used to visualize, record, and classify plankton. Its modular design and open hardware approach make it a cost-effective alternative to more expensive systems like the FlowCam. Depending on whether you build it yourself from components or purchase a fully assembled unit, costs can range from about $500 to $9,000. Despite the lower price point, the PlanktoScope can achieve high image resolution and successful cell quantification, making it a powerful tool for both research and education. Its portability and easily replaceable parts make the PlanktoScope especially valuable for fieldwork and citizen science initiatives.
This summer, I’ve focused on optimizing the PlanktoScope settings to match the phytoplankton cell counts typically obtained from the FlowCam, which will be taken on a research cruise in August. Elevated sea surface temperatures in the late summer are expected to drive blooms of Akashiwo sanguinea, a red-tide forming organism of concern. These blooms have been linked to oyster larvae mortality events in the Skidaway River Estuary, highlighting the need for accurate plankton imaging and counting. Real-time monitoring with tools like the PlanktoScope and FlowCam can help inform local aquaculture stakeholders and support the Ph.D. research of Mallory Mintz, who is investigating the physiochemical drivers behind Akashiwo bloom dynamics.
Mary with the FlowCam, which will be gone for a cruise in the Gulf :(
Summer Internship Project & Experiences
I began the summer by calibrating the PlanktoScope to improve its performance and image quality. This involved adjusting the auto white balance, optimizing pump efficiency, and refining particle size measurements. I also spent time learning how to focus the camera, using a combination of microbeads and larger plankton samples from net tows to maximize the number of focused cells per image.
Assembled Planktoscope
After setup optimization, I began running trials to compare Akashiwo sanguinea counts between the PlanktoScope and the FlowCam. I started with a concentrated culture to provide a clear and quantifiable reference for comparison. FlowCam analysis measured the concentration at 503.84 cells/mL. By matching the imaged fluid volume exactly on the PlanktoScope, I obtained a comparable result of 478.58 cells/mL after image processing. This level of variation falls well within the expected range for repeated FlowCam runs on the same sample, which was a very encouraging outcome. However, I quickly found the PlanktoScope’s built-in image processing to not be practical for daily use. Although it is effective at cleaning the background and isolating regions of interest, about 95% of the detected objects were bubbles or debris, with only 5% being actual phytoplankton cells.
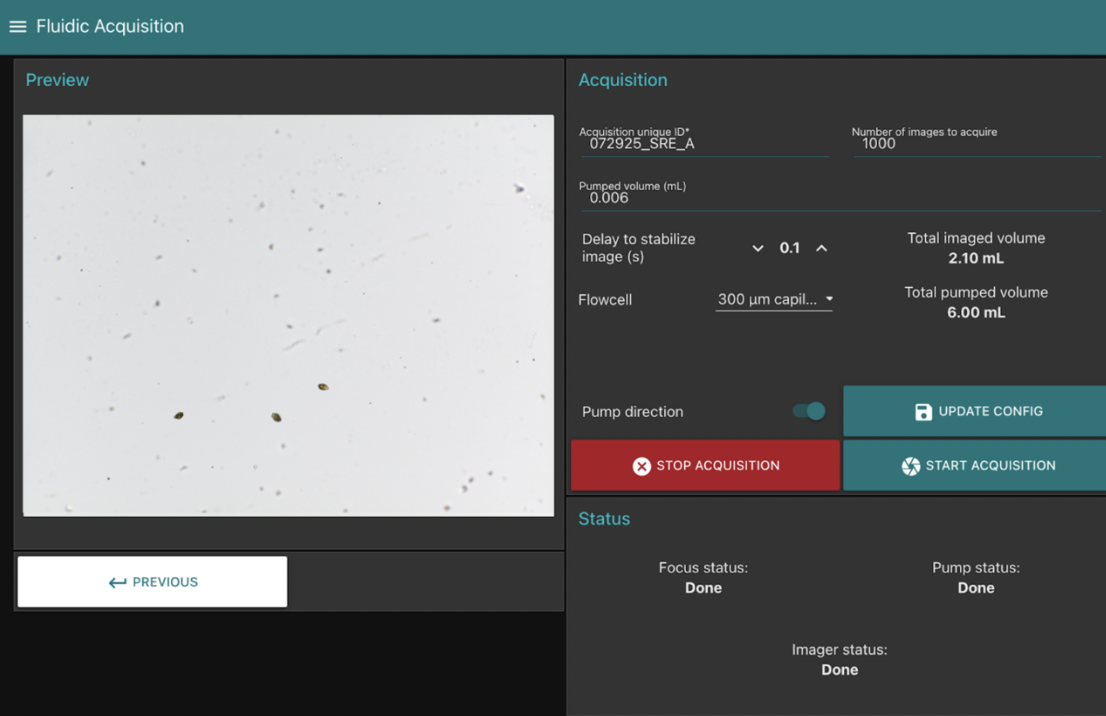
User Interface & Example Settings for Estuary Run
To improve image analysis, Mallory Mintz adapted a workflow from Dr. Adam Greer’s lab that utilizes flatfielding and ImageJ. Flatfielding removes background noise by averaging 100 images and subtracting that average from each frame. This successfully highlights the phytoplankton, and ImageJ can then isolate regions of interest for counting.
After multiple intercomparison trials, I found that collecting around 1000 images per run creates a strong match to FlowCam data while keeping the file size manageable for daily processing. We have now moved on to using real estuary water, which has produced comparable results. We are continuing to fine-tune the image analysis settings to detect additional phytoplankton species alongside A. sanguinea, however we are confident that the PlanktoScope can serve as a worthy substitute for cell counts while the FlowCam is away.
Estuary net tow photo of community
Throughout this work, I have followed a protocol written by PlanktoScope co-developer Fabien Lombard, which has been very helpful. As I gained experience, I modified the protocol to better suit our lab’s needs, particularly for harmful algal bloom monitoring. As my internship concludes, I hope the PlanktoScope remains a useful tool for future students, public outreach events, and field research opportunities once the FlowCam returns.
Akashiwo images from the Planktoscope:
PlanktoScope Community
One of the most enjoyable aspects of working with the PlanktoScope has been its open-source and community-driven nature. All software versions are publicly available on GitHub, where developers actively seek feedback and contributions from users. There is also an active PlanktoScope Slack community, with conversations ranging from hardware assembly tips to advice on taking the instrument through TSA. The developers themselves are highly engaged in these discussions and have been an invaluable resource as I’ve worked through various technical challenges. This strong sense of collaboration has made learning and troubleshooting with the PlanktoScope very enriching.